I wrote a very simple and user-friendly method, that I called ddeint, to solve delay differential equations (DDEs) in Python, using the ODE solving capabilities of the Python package Scipy. As usual the code is available at the end of the post :).
Example 1 : Sine
I start with an example whose exact solution is known so that I can check that the algorithm works as expected. We consider the following equation:
The trick here is that so the exact solution of this equation is actually the sine function.
from pylab import * model = lambda Y,t : Y(t - 3*pi/2) # Model tt = linspace(0,50,10000) # Time start, end, and number of points/steps g=sin # Expression of Y(t) before the integration interval yy = ddeint(model,g,tt) # Solving # PLOTTING fig,ax=subplots(1) ax.plot(tt,yy,c='r',label="$y(t)$") ax.plot(tt,sin(tt),c='b',label="$sin(t)$") ax.set_ylim(ymax=2) # make room for the legend ax.legend() show()
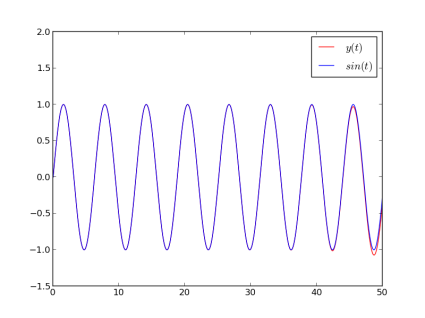
The resulting plot compares our solution (red) with the exact solution (blue). See how our result eventually detaches itself from the actual solution as a consequence of many successive approximations ? As DDEs tend to create chaotic behaviors, you can expect the error to explode very fast. As I am no DDE expert, I would recommend checking for convergence in all cases, i.e. increasing the time resolution and see how it affects the result. Keep in mind that the past values of Y(t) are computed by interpolating the values of Y found at the previous integration points, so the more points you ask for, the more precise your result.
Example 2 : Delayed negative feedback
You can select the parameters of your model at integration time, like in Scipy’s ODE and odeint. As an example, imagine a product with degradation rate r, and whose production rate is negatively linked to the quantity of this same product at the time (t-d):
We have three parameters that we can choose freely. For K = 0.1, d = 5, r = 1, we obtain oscillations !
from pylab import * # MODEL, WITH UNKNOWN PARAMETERS model = lambda Y,t,k,d,r : 1/(1+(Y(t-d)/k)**2) - r*Y(t) # HISTORY g = lambda t:0 # SOLVING tt = linspace(0,50,10000) yy = ddeint(model,g,tt,fargs=( 0.1 , 5 , 1 )) # K = 0.1, d = 5, r = 1 # PLOTTING fig,ax=subplots(1) ax.plot(tt,yy,lw=2) show()
Example 3 : Lotka-Volterra system with delay
The variable Y can be a vector, which means that you can solve DDE systems of several variables. Here is a version of the famous Lotka-Volterra two-variables system, where we introduce some delay d. For d=0 we find the solution of a classical Lotka-Volterra system, and for d non-nul, the system undergoes an important amplification:
from pylab import *
def model(Y,t,d):
x,y = Y(t)
xd,yd = Y(t-d)
return array([0.5*x*(1-yd), -0.5*y*(1-xd)])
g = lambda t : array([1,2])
tt = linspace(2,30,20000)
fig,ax=subplots(1)
for d in [0, 0.2]:
yy = ddeint(model,g,tt,fargs=(d,))
# WE PLOT X AGAINST Y
ax.plot(yy[:,0],yy[:,1],lw=2,label='delay = %.01f'%d)
ax.legend()
show()
Example 4 : A DDE with varying delay
This time the delay depends on the value of Y(t) !
from pylab import * model = lambda Y,t: -Y(t-3*cos(Y(t))**2) tt = linspace(0,30,2000) yy = ddeint(model, lambda t:1, tt) fig,ax=subplots(1) ax.plot(tt,yy,lw=2) show()
Code
Explanations
The code is written on top of Scipy’s ‘ode’ (ordinary differential equation) class, which accepts differential equations under the form
model(Y,t) = ” expression of Y'(t) ”
where , and the output
, must be Numpy arrays (i.e. vectors).
For our needs, we need the input to be a function of time, more precisely a function that can compute
at any past or present
using the values of
already computed. We also need
to return the value of some function
if t is inferior to some time
that marks the start of the integration.
To this end, I first implemented a class (ddeVar) of variables/functions which can be called at any time : for $t<latex t_c$, it will return the value of
, and for
, it will look for two already computed values
and
at times
, from which it will deduce
using a linear interpolation. Scipy offers many other kinds of interpolation, but these will be slower and won't support vectors for
.
Such variables need to be updated every time a new value of is computed, so I created a class 'dde' that inherits from Scipy's 'ode' class but overwrites its integration method so that our special function
is updated after each integration step. Since 'ode' would feed the model with a vector Y (a Numpy array to be precise), which we don't want, we give to the integrator an interface function that takes a Numpy array Y as an argument, but immediately dumps it and calls the model with our special ddeVar variable Y (I hope that was clear 🙂 ).
Ok, here you are for the code
You will find the code and all the examples as an IPython notebook HERE (if you are a scientific pythonist and you don’t know about the IPython notebook, you are really missing something !). Just change the extension to .ipynb to be able to open it. In case you just asked for the code:
# REQUIRES PACKAGES Numpy AND Scipy INSTALLED
import numpy as np
import scipy.integrate
import scipy.interpolate
class ddeVar:
""" special function-like variables for the integration of DDEs """
def __init__(self,g,tc=0):
""" g(t) = expression of Y(t) for t<tc """
self.g = g
self.tc= tc
# We must fill the interpolator with 2 points minimum
self.itpr = scipy.interpolate.interp1d(
np.array([tc-1,tc]), # X
np.array([self.g(tc),self.g(tc)]).T, # Y
kind='linear', bounds_error=False,
fill_value = self.g(tc))
def update(self,t,Y):
""" Add one new (ti,yi) to the interpolator """
self.itpr.x = np.hstack([self.itpr.x, [t]])
Y2 = Y if (Y.size==1) else np.array([Y]).T
self.itpr.y = np.hstack([self.itpr.y, Y2])
self.itpr.fill_value = Y
def __call__(self,t=0):
""" Y(t) will return the instance's value at time t """
return (self.g(t) if (t<=self.tc) else self.itpr(t))
class dde(scipy.integrate.ode):
""" Overwrites a few functions of scipy.integrate.ode"""
def __init__(self,f,jac=None):
def f2(t,y,args):
return f(self.Y,t,*args)
scipy.integrate.ode.__init__(self,f2,jac)
self.set_f_params(None)
def integrate(self, t, step=0, relax=0):
scipy.integrate.ode.integrate(self,t,step,relax)
self.Y.update(self.t,self.y)
return self.y
def set_initial_value(self,Y):
self.Y = Y #!!! Y will be modified during integration
scipy.integrate.ode.set_initial_value(self, Y(Y.tc), Y.tc)
def ddeint(func,g,tt,fargs=None):
""" similar to scipy.integrate.odeint. Solves the DDE system
defined by func at the times tt with 'history function' g
and potential additional arguments for the model, fargs
"""
dde_ = dde(func)
dde_.set_initial_value(ddeVar(g,tt[0]))
dde_.set_f_params(fargs if fargs else [])
return np.array([g(tt[0])]+[dde_.integrate(dde_.t + dt)
for dt in np.diff(tt)])
Other implementations
If you need a faster or more reliable implementation, have a look at the packages pyDDE and pydelay, which seem both very serious but are less friendly in their syntax.